Transcription factors bind to cis-regulatory elements (transcription factor binding site, TFBS) to regulate the transcription of the downstream gene (Latchman, 1997). Variants within TFBSs can impact the binding strength of transcription factors and participate in the biogenesis of human diseases, including cancers (Huang et al., 2014; Liu et al., 2017). Although many tools have been used to evaluate the functional effects of these TFBS variants (Boyle et al., 2012; Coetzee et al., 2015; Fu et al., 2014; Kumar et al., 2017; Ward and Kellis, 2016; Zuo et al., 2015), these tools generally handle each variant independently, neglecting the potential “interference” resulting from multiple variants within the same TFBS.
To handle the compound effects within TFBSs, we developed COPE-TFBS by comparing the position weight matrix score of the mutant and wild-type TFBS sequences. To the best of our knowledge, COPE-TFBS is the first tool that annotates the variant effects on TFBSs by considering the entire sequence context. Applying COPE-TFBS to thousands of human genomes, 1,502 emerging novel TFBS, 266 transformed TFBS (a known TFBS is transformed into another TFBS) as well as more than 85,000 discordantly annotated TFBS (a variant could increase the binding strength of transcription factor and another variant could decrease the binding strength) cases were identified after scanning data from 1000 Genomes and GTEx Projects with COPE-TFBS. Furthermore, these compound effects could affect eQTLs, the expression of downstream genes and disease-associated regulatory variants. Together with COPE-PCG (Cheng et al., 2017), this work not only demonstrates the importance of the long-neglected compound effects, but also offers a unique toolkit for the community.
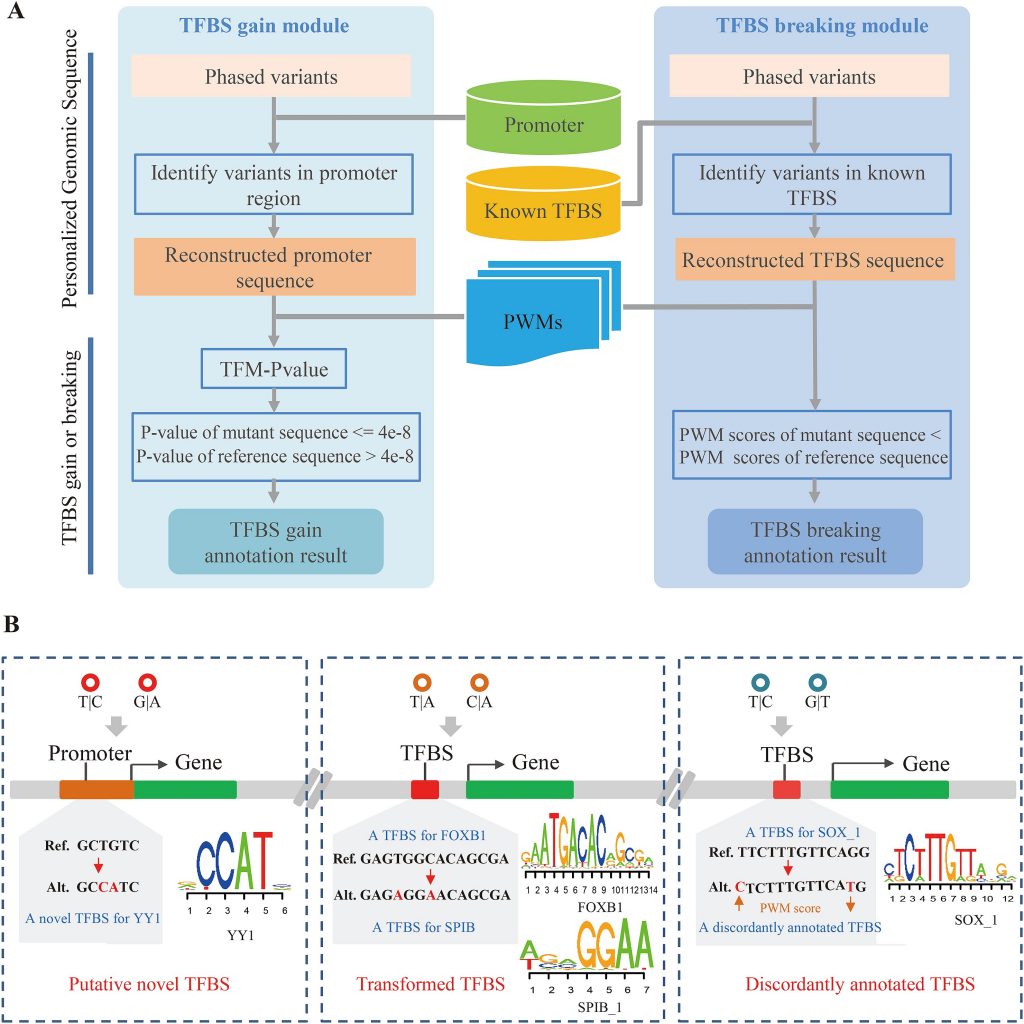
(A) The workflow of COPE-TFBS. (B) The compound effect resulting from multiple variants.
The paper entitled “Systematic identification and annotation of multiple-variant compound effects at transcription factor binding sites in human genome” is online at Journal of Genetics and Genomics (JGG), with the Web server available publically at http://cope.gao-lab.org
Website: http://cope.gao-lab.org
Paper link: https://doi.org/10.1016/j.jgg.2018.05.005
Reference:
Cheng, S.J., Shi, F.Y., Liu, H., Ding, Y., Jiang, S., Liang, N., Gao, G., 2017. Accurately annotate compound effects of genetic variants using a context-sensitive framework. Nucleic acids research 45, e82.
Latchman, D.S., 1997. Transcription factors: an overview. Int J Biochem Cell Biol 29, 1305-1312.
Huang, Q., Whitington, T., Gao, P., Lindberg, J.F., Yang, Y., Sun, J., Vaisanen, M.R., Szulkin, R., Annala, M., Yan, J., Egevad, L.A., Zhang, K., Lin, R., Jolma, A., Nykter, M., Manninen, A., Wiklund, F., Vaarala, M.H., Visakorpi, T., Xu, J., Taipale, J., Wei, G.H., 2014. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nature genetics 46, 126-135.
Liu, N.Q., Ter Huurne, M., Nguyen, L.N., Peng, T., Wang, S.Y., Studd, J.B., Joshi, O., Ongen, H., Bramsen, J.B., Yan, J., Andersen, C.L., Taipale, J., Dermitzakis, E.T., Houlston, R.S., Hubner, N.C., Stunnenberg, H.G., 2017. The non-coding variant rs1800734 enhances DCLK3 expression through long-range interaction and promotes colorectal cancer progression. Nature communications 8, 14418.
Boyle, A.P., Hong, E.L., Hariharan, M., Cheng, Y., Schaub, M.A., Kasowski, M., Karczewski, K.J., Park, J., Hitz, B.C., Weng, S., Cherry, J.M., Snyder, M., 2012. Annotation of functional variation in personal genomes using RegulomeDB. Genome research 22, 1790-1797.
Coetzee, S.G., Coetzee, G.A., Hazelett, D.J., 2015. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics 31, 3847-3849.
Fu, Y., Liu, Z., Lou, S., Bedford, J., Mu, X.J., Yip, K.Y., Khurana, E., Gerstein, M., 2014. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome biology 15, 480.
Kumar, S., Ambrosini, G., Bucher, P., 2017. SNP2TFBS – a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic acids research 45, D139-D144.