The Cell BLAST study (Searching large-scale scRNA-seq databases via unbiased cell embedding with Cell BLAST) was recently awarded by Genomics, Proteomics and Bioinformatics (GPB) as China’s top ten bioinformatics advances of 2020.
As a powerful tool for studying cellular heterogeneity, single-cell transcriptomic sequencing has seen rapid development in recent years, with large amounts of data continuously being accumulated. To better utilize these valuable data, we developed a single-cell transcriptomic data integration and querying method called Cell BLAST. Analogous to the BLAST algorithm for studying biological sequences, Cell BLAST can query and annotate newly-generated single-cell data in an existing database with high efficiency and accuracy. Not only can it save the time of manual annotation based on known marker genes, but also reduce the liability of human error. By employing adversarial learning, Cell BLAST effectively addresses the problem of multi-level batch effect in single-cell transcriptomic querying. Moreover, based on the characterization of intrinsic stochasticity in single-cell measurements, Cell BLAST proposes a new cell-to-cell similarity metric called NPD. Combination of the two enables effective data integration and comparative analysis across multiple single-cell datasets. To make better use of the capabilities of Cell BLAST, we further constructed a single-cell transcriptomic reference databased called ACA, covering a wide variety of tissues and organs across diverse species, and provide a Web-based online querying service (https://cblast.gao-lab.org). This work provides new tool and resource for cell annotation and cross-dataset analysis based on the effective utilization of existing data. Meanwhile, it also demonstrates the pivotal role of computational biology and bioinformatics in studying complex biological systems.
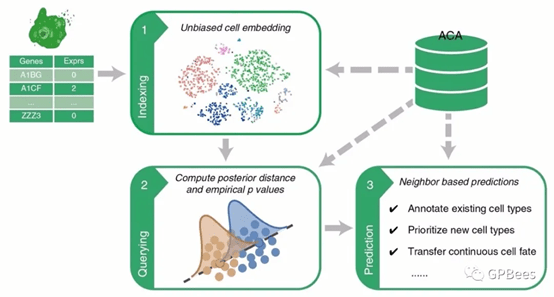
Figure. Flowchart of single-cell transcriptomic querying method Cell BLAST
Cell BLAST first projects the query data and ACA reference data to a low-dimensional cell embedding space, where adversarial learning is employed to remove multi-level batch effect. Based on the characterization of intrinsic stochasticity of single-cell measurements, it further uses the NPD metric to identify reference cells most similar to the query data, which are then used for automatic data annotation.
The “China’s top ten bioinformatics” series was initiated by Genomics, Proteomics and Bioinformatics (GPB) from 2018, aimed at promoting innovation and showcasing major advances in China’s bioinformatics research. Previously, the human lncRNA study from Gao lab was awarded as China’s top ten bioinformatics database of 2019.
Application and database URL:
Publication:
Cao, Z. J., Wei, L., Lu, S., Yang, D. C. & Gao, G. Searching large-scale scRNA-seq databases via unbiased cell embedding with Cell BLAST. Nat. Commun. 2020; 11:3458. PMID: 32651388